
How knowledge graph technology is helping Cochrane respond to COVID-19
Evidence-Based
Many will have heard of Cochrane, maybe more now than ever before. Cochrane is the fabulous global not-for-profit organisation whose core mission is to put evidence at the heart of health decision-making all over the world.
Data Language has been a technology partner of Cochrane for more than 6 years now. Over this time we have helped Cochrane implement a radical new data architecture centred around linked data and knowledge graph technologies improving the mechanisms through which Cochrane describe evidence in healthcare using structured data.
One of Cochrane’s primary outputs is the production of `systematic reviews`. A typical Cochrane review is a meta-analysis performed over a set of closely related clinical trials that establishes the risk-of-bias in those studies. Producing the systematic review is a complex, domain specific, highly knowledge-bound task that can take months for subject matter experts (typically researchers, clinicians, and health professionals) to complete. The completed review itself is a complex semi-structured document, a compilation of the evidence, the input data, the analytics, and the conclusions of the Cochrane review group. The review is the gold-standard in healthcare evidence. It really is.
While there is an awful lot we can talk about here, I want to focus on how we have been able to rapidly adapt the Cochrane technical and data architecture to deploy an app for collating and navigating COVID-19 living evidence in super-rapid time: the Cochrane COVID-19 Study Register.
The key to this, has been a flexible (knowledge) graph based linked data architecture, micro-services and full automation of the technical stack. My colleague Julian Everett provides further background and insight into the technical and data strategy at Cochrane here.
Information Architecture
Our initial explorations into the use of linked data at Cochrane began some 8 years ago, when a conversation over a beer between Silver Oliver (the Data Language head of Information Architecture), and Chris Mavergames (now the Cochrane CIO) that led us to build a prototype exploring how linked data could be used to augment Cochrane reviews and the source studies with structured data, to enable Cochrane to interrogate their data in different and interesting ways.
The principle here was one of annotation and augmentation. We could use a graph database (a knowledge graph), to capture evidential statements about the content and describe the evidence at the right places within the content using structured linked data.
This required :
- A model for describing clinical questions (the evidence)
- A linked data vocabulary that we could use to describe and construct the clinical questions
- A content model - describing where in the content (the studies and reviews) we need to make structured data evidence annotations
- An annotation model that captured the provenance and workflow of the curation of the evidence assertions.
- A knowledge graph implementation to store the linked data vocabulary, the content metadata, and the evidence.
- Tools to enable this augmentation
We essentially adopted what at the time was a burgeoning pattern for the semantic annotation of content, that myself and Silver and others had been working on at the BBC. For Cochane this pattern comprised :
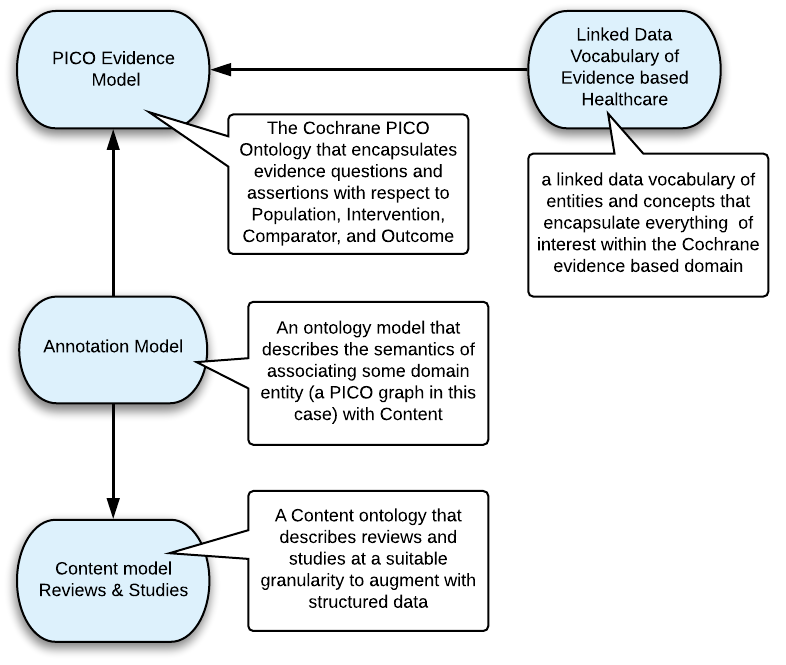
PICO - Describing the Evidence
The cornerstone of the pattern is the ontology model for describing clinical questions, or evidence. Working with Cochrane subject matter experts we designed the PICO Ontology. The acronym comprises :
P - Population, describing the demography of the subjects being studied, the ages, sex, the condition(s), existing treatments, and the social context.
I - an Intervention being studied - including surgical procedures, psychotherapies, educational interventions or even health system changes. Combinations of interventions can be represented, as well as drugs and their dosage and frequency, setting, mode of delivery, and also devices used in delivering the intervention.
C - Comparison - another intervention that is being compared with the primary intervention under study, including placebos.
O - the Outcome for the population being studied, including subsequent conditions, treatments, metrics and outcome classification.
The PICO ontology model has since become widely adopted for describing healthcare evidence, furthermore is equally applicable in other evidence-based domains. The ontology has evolved through a number of iterations since inception to encapsulate complex graph representations of evidence. The latest PICO ontology conforms to the principles of RDF and OWL modelling, and is depicted as follows, where visualisation shows Classes as blue ovals, and properties as yellow hexagons :
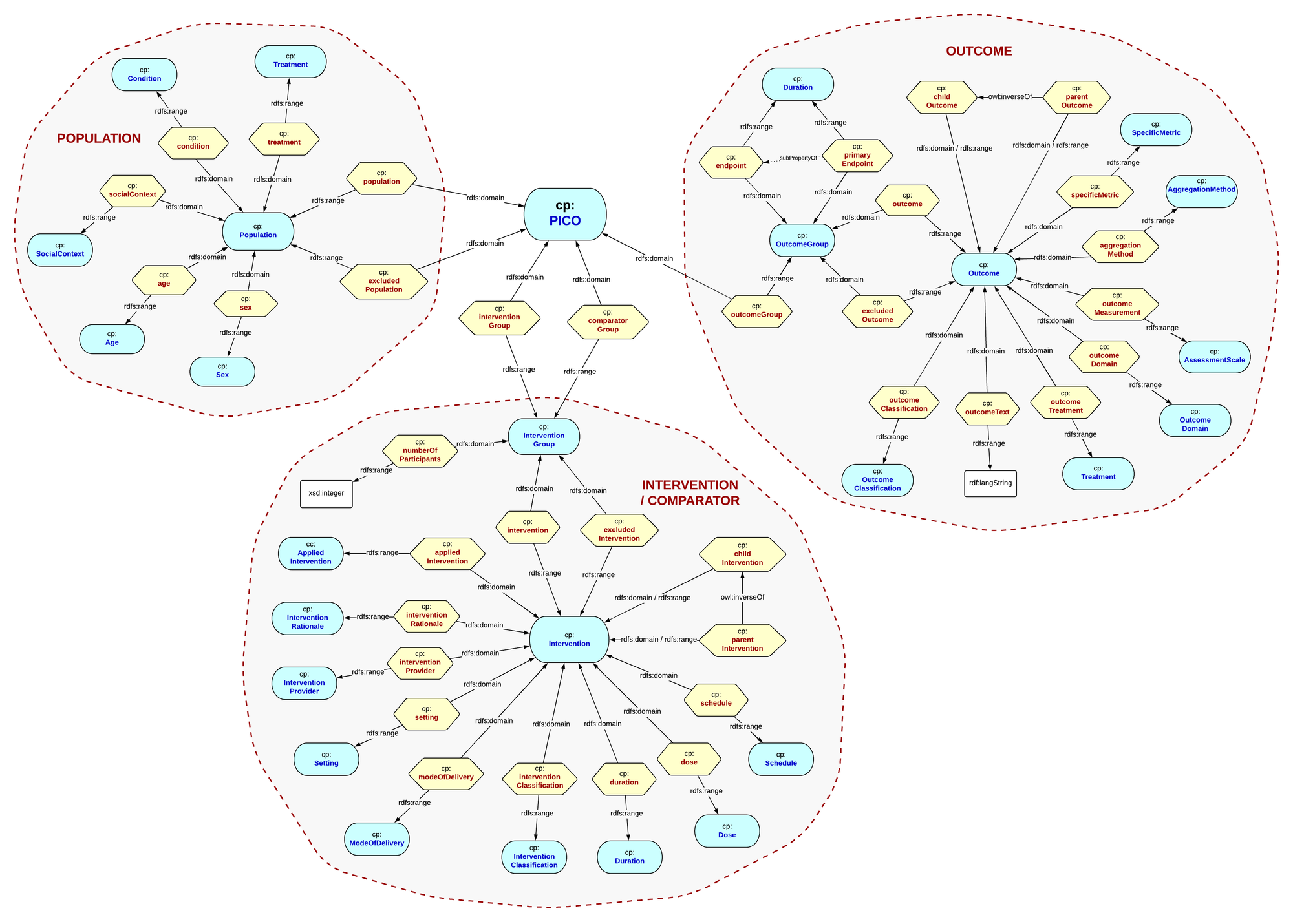
We can then construct PICO instances conforming to the ontology model. The model allows us to specify complex populations; populations including some demographic, and excluding others. It lets us specify detailed interventions that include combinations of drugs and therapies, with different dosages, schedule and duration. We can construct parent/child and grouped interventions. Similarly outcomes can be excluded, and arranged in parent/child relationships. What we have constructed is a rich PICO graph that describes a specific evidential case. The entity instances assigned to leaf nodes on each PICO graph are drawn from the Cochrane Linked Data Vocabulary - instances of drugs, procedures etc (more on this later).
Describing the Content
Similarly the content model is an important part of the puzzle. We treat this as a separate bounded context. Our content model is specific to the Cochrane domain. If you were annotating PICOs against your own (different) content, then you would need a content model that specifically described that. Within the Cochrane domain we are interested in Studies, Reviews, and the subcomponents of Reviews that we need to augment with PICOs, for example the meta-analysis, and each included Study that was reviewed. The Content model looks like this :
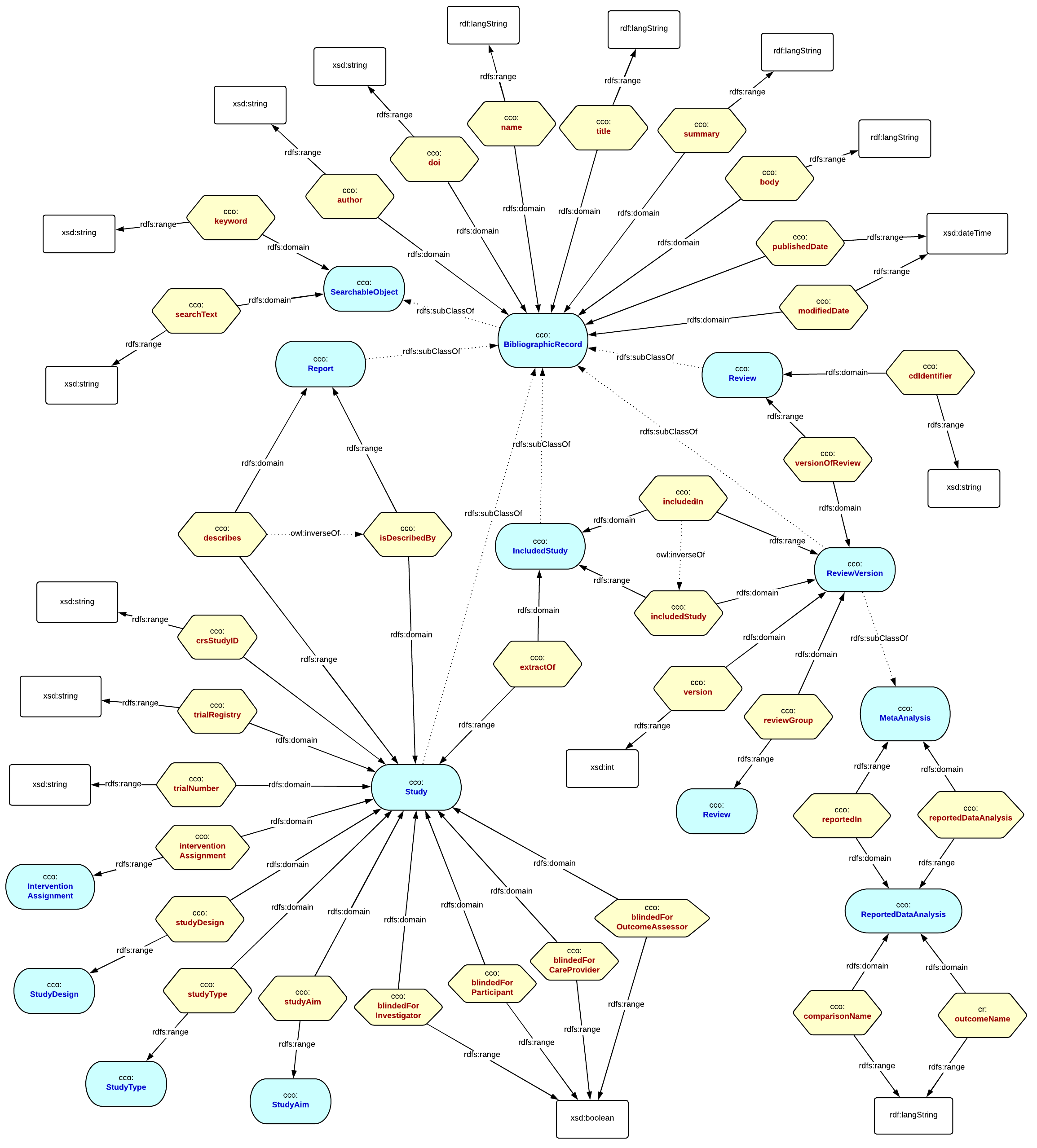
Semantic Annotation
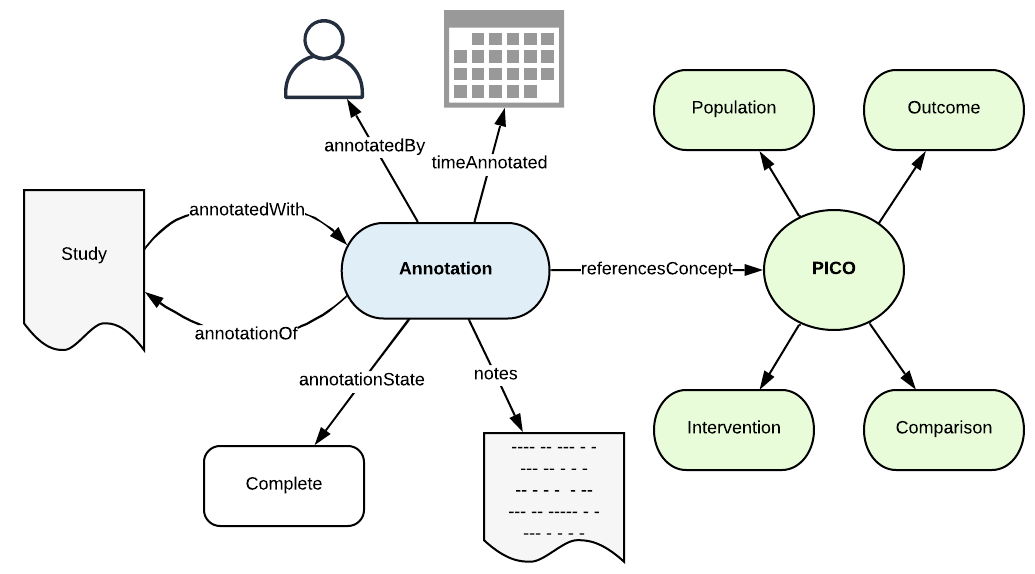
We now need a mechanism to augment the content with PICOs and capture the provenance of the augmentation. We call this semantic annotation and again this has an annotation ontology that describes the metadata we need to capture.
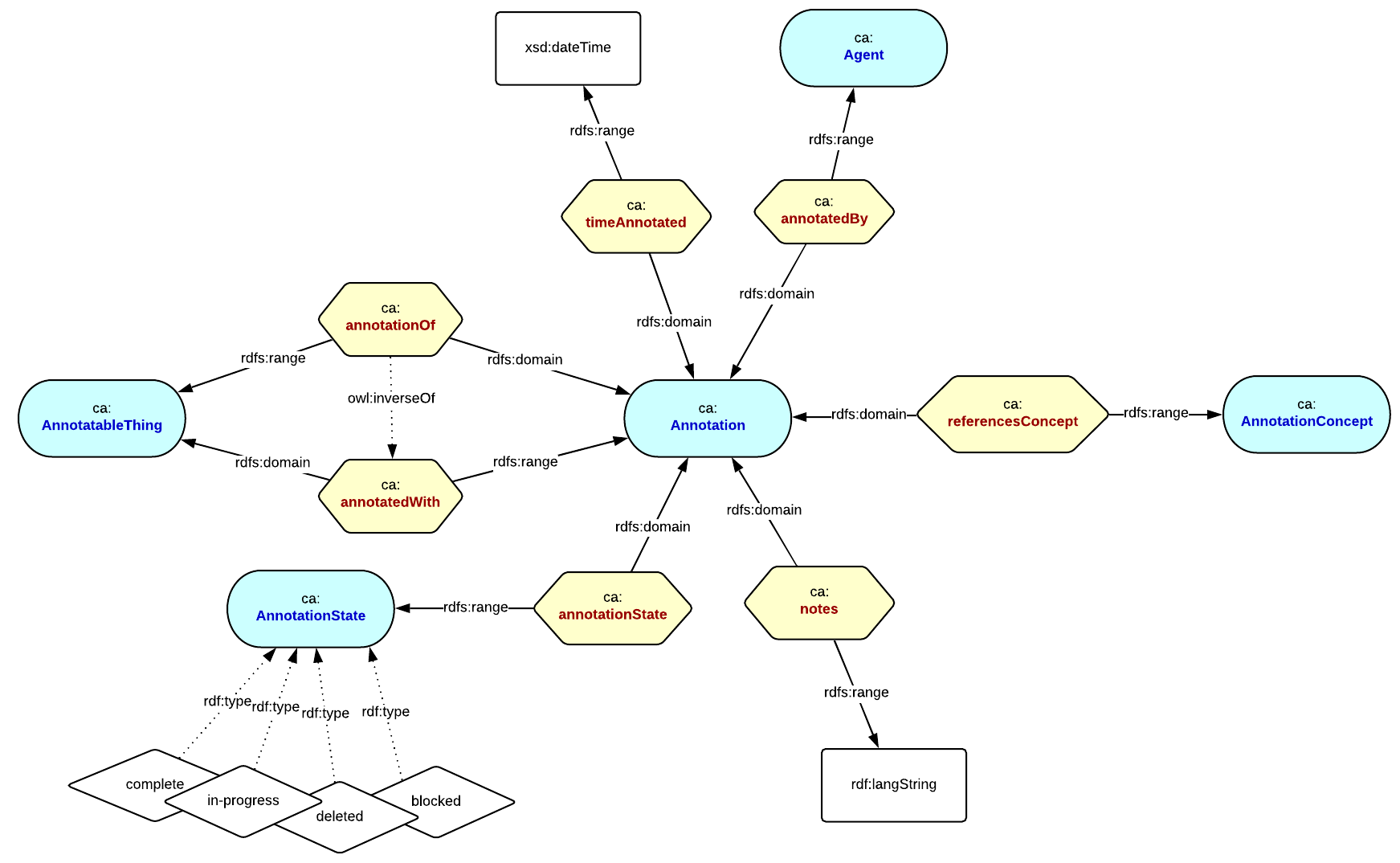
This is a bounded context ontology that can be used for annotating anything annotatable (an AnnotatableThing) that can be referenced by an IRI with some resource or concept. In this case, we are annotating entities in the Cochrane content model, with PICOs. The `Annotation` captures the workflow and provenance metadata resulting from the act of making an annotation, for example:
The Cochrane Linked Data Vocabulary
The Cochrane linked data vocabulary consists of approximately 400k linked data terms that Cochrane uses to describe the domain of evidence based health care. We use our linked data to construct clinical questions using the PICO Graph Ontology
The concepts in the vocabulary include the terms that describe the Populations, Interventions, Conditions, and Outcomes, and are ultimately used to construct PICO graphs themselves describing the evidence within Cochrane Reviews and Studies.
The Cochrane Vocabulary, where possible, links to existing health vocabularies including MeSH, SNOMED-CT, MedDRA, RxNorm, and ATC, thereby supporting data reuse and semantic standardization across health and social care informatics. While the vocabulary looks taxonomical in nature, an individual concept within it may have multiple parent concepts. Its structure is thus a directed graph (digraph).
Each concept in the vocabulary is represented in RDF, and is available via API in JSON-LD format, for example, the COVID-19 Condition is:
{
"id": "http://data.cochrane.org/concepts/NrO30O5ZnYIkjE",
"type": "http://data.cochrane.org/ontologies/core/Condition",
"label": "COVID-19",
"lastModifiedDate": "2020-03-19T10:05:28.566Z",
"broaderConcept": [
"http://data.cochrane.org/concepts/r4hp38cvdb8m"
],
"alternateLabel": [
"Coronavirus disease 2019",
"2019-nCoV acute respiratory disease",
"Novel coronavirus pneumonia",
"Wuhan flu",
"Wuhan pneumonia"
],
"@context": "/concepts/context"
}The ontology model for the vocabulary can be seen on the homepage of the linked data vocabulary browser.
The Knowledge Graph
At the core of our data architecture is a knowledge graph. The ontology models, the vocabulary, the content metadata, and the PICOs are all stored in the knowledge graph. The physical manifestation of this is an RDF compliant graph database, and in this case we are using Ontotext’s GraphDB. The knowledge graph lets us ask questions of our data using the W3C SPARQL query language. Specifically, we can query our content via the PICO graphs and Cochrane vocabulary terms. Now we have clinical questions described using a shared vocabulary with structured semantics we can leverage it to aggregate studies, reviews and meta-analyses with similar PICO fingerprints, or find patterns in the PICOs (the clinical questions) themselves.
Moreover, we can use the RDFS and OWL reasoning supported in the graph database, to leverage inferences such as transitivity across the linked data vocabulary, for example a query for studies with populations having the Condition `Coronavirus infection` (`https://data.cochrane.org/concepts/r4hp38cvdb8m`) would surface those with Conditions including MERS, SARS, and COVID-19. Even this fairly light use of semantic reasoning, allows us to build some high-utility user-facing applications, including the COVID-19 Study Register described here.
Services
To make all this a reality, we needed to integrate the curation of the linked data vocabulary and the PICO Graphs themselves into the Cochrane SME’s and editor’s daily workflows. We needed tools. To build tools we need data services. Enter ElasticSearch, NodeJS, Docker and Kubernetes brought together in the following conceptual data architecture :
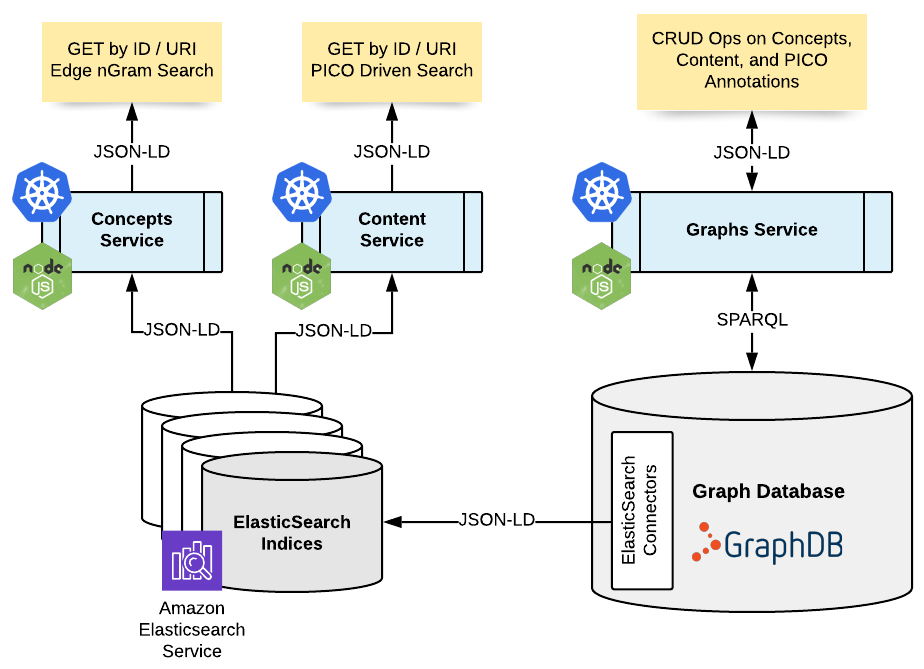
A key pattern we are using here is the combination of GraphDB with ElasticSearch. One of the unique (killer) features of GraphDB is its ElasticSearch Connector plugin. These connectors allow you to connect the graph database to an elasticsearch cluster, and materialise subgraphs into an elasticsearch index as structured JSON documents or even fully formed JSON-LD entities. A connector is defined in the form of a SPARQL update providing a mapping between graph property chains, and the JSON/JSON-LD output required. Once a connector has been configured, as RDF statements are written into the graph, if the statements match the mappings defined in the connector, the corresponding JSON document in the elasticsearch index is inserted or modified accordingly. This synchronisation between the graph and the elastic index, all happens post-inference, and within the write transaction of the data into the graph.
It is hard to put into a couple of paragraphs how flexible and powerful this feature is, but it essentially allows you to combine all the benefits of write time RDFS/OWL reasoning in the graph, with offloaded highly scalable, rocket fast reads and search of post-inference entities in Elastic. All the synchronization of data between the two is maintained by the connector.
Once these connectors are in place, all that is left is to develop and deploy microservices to interface to both the graph (for CRUD operations), and ElasticSearch for lookup and search operations. These were built in NodeJS (as we are consuming and producing JSON / JSON-LD NodeJS is the ideal choice for efficient coding of these services), containerised with Docker and deployed into an AWS EKS Kubernetes cluster. The build, test, and deployment of the services are fully automated using Terraform, Helm, and continuous deployment pipelines.
Tooling
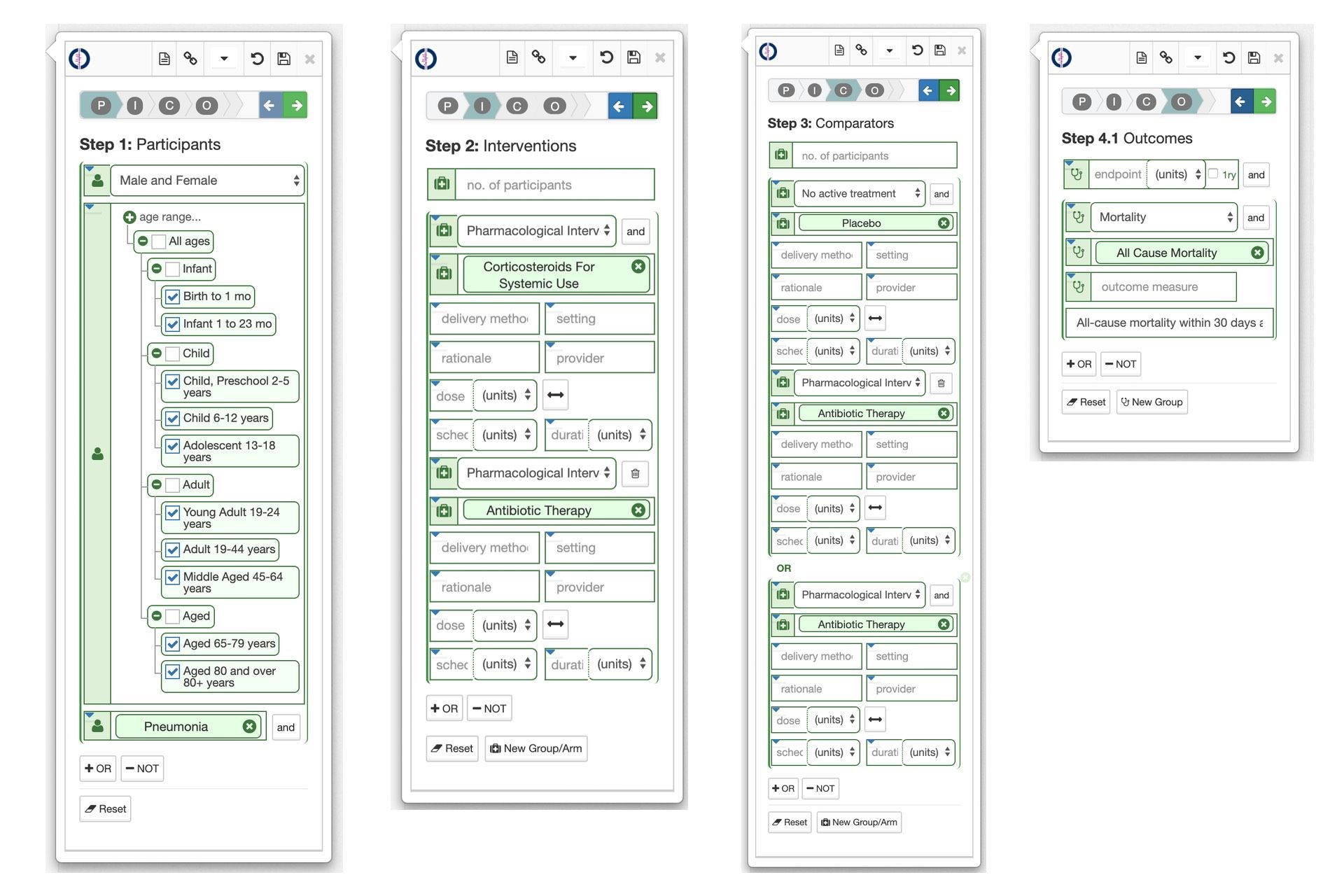
Building upon the microservices we needed tools that could be integrated into the Cochrane editor workflows, for curating, moderating, and performing quality assurance on the PICOs being curated. For curation the approach taken was to build a PICO Annotator widget that could be dropped into existing Cochrane web-base editing tools, or used standalone.
This PICO Annotator widget allows users to curate a PICO in-situ using a simple step-by-step wizard-based user experience. Each step in the widget lets the user construct one of the P, I, C, O components of the PICO graph as follows, leaning heavily on the controlled linked data vocabularies in autocomplete and drop down controls :
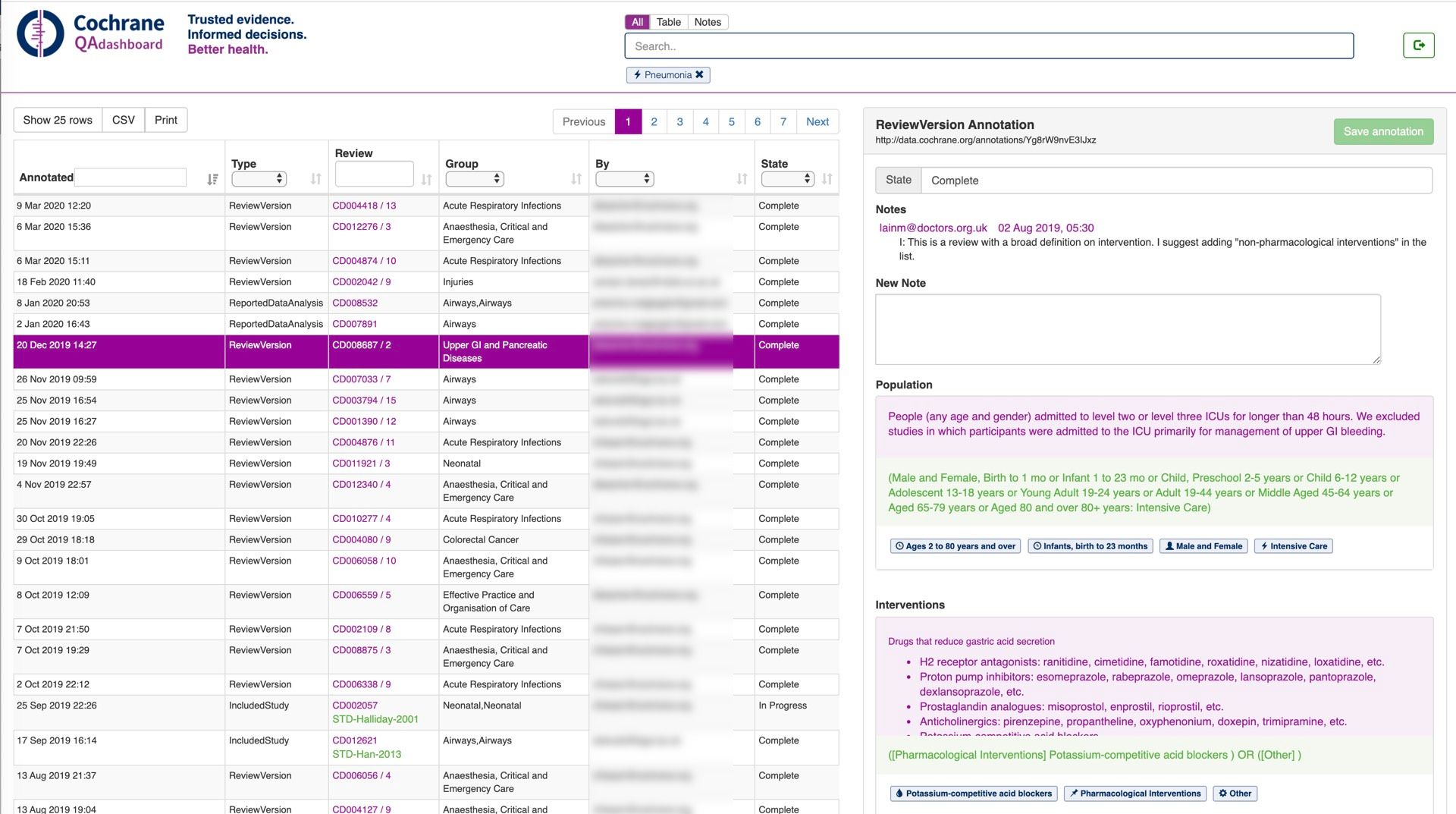
To provide Quality Assurance on PICOs curated we built a QA Dashboard, that provided features for subject matter experts to review, comment on, ask questions about, and update workflow statuses of PICO annotations.
The result of this process is a PICO Graph annotated at a specific place in a Review or Study conforming to the PICO Ontology. In JSON-LD form it looks like this :
{
"id": "http://data.cochrane.org/picos/ADeP1e0YdNIynG",
"type": "PICO",
"population": {
"id": "http://data.cochrane.org/populations/GEqPBqRaXYhP2W",
"type": "Population",
"age": [
{
"id": "http://data.cochrane.org/concepts/kk05h7rpym8z",
"type": "Age",
"label": "Adult 19-44 years"
},
{
"id": "http://data.cochrane.org/concepts/kk05h7rpym90",
"type": "Age",
"label": "Middle Aged 45-64 years"
},
{
"id": "http://data.cochrane.org/concepts/kk05h7rpym91",
"type": "Age",
"label": "Aged 65-79 years"
},
{
"id": "http://data.cochrane.org/concepts/kk05h7rpym92",
"type": "Age",
"label": "Aged 80 and over 80+ years"
}
],
"condition": {
"id": "http://data.cochrane.org/concepts/r4hp3p86xjng",
"type": "Condition",
"label": "Pneumonia"
},
"sex": {
"id": "http://data.cochrane.org/concepts/lr5qxyw6ww35",
"type": "Sex",
"label": "Male and Female"
}
},
"interventionGroup": {
"id": "http://data.cochrane.org/intervention-groups/PNJK3JWRZYfWM4",
"type": "InterventionGroup",
"intervention": {
"id": "http://data.cochrane.org/interventions/q06N36okevhAmD",
"type": "Intervention",
"childIntervention": [
{
"id": "http://data.cochrane.org/interventions/916PZ6BOMqfOrm",
"type": "Intervention",
"appliedIntervention": {
"id": "http://data.cochrane.org/concepts/r4hp13mgns50",
"type": "DrugCategory",
"label": "Corticosteroids For Systemic Use"
},
"interventionClassification": {
"id": "http://data.cochrane.org/concepts/kn3ptfq7c6lz",
"type": "InterventionClassification",
"label": "Pharmacological Interventions"
}
},
{
"id": "http://data.cochrane.org/interventions/Yg4nv4MmpYI4P3",
"type": "Intervention",
"appliedIntervention": {
"id": "http://data.cochrane.org/concepts/r4hp5zcz3yj6",
"type": "Procedure",
"label": "Antibiotic Therapy"
},
"interventionClassification": {
"id": "http://data.cochrane.org/concepts/kn3ptfq7c6lz",
"type": "InterventionClassification",
"label": "Pharmacological Interventions"
}
}
]
}
},
"comparatorGroup": {
"id": "http://data.cochrane.org/intervention-groups/j0Z2DZEkX6h3JD",
"type": "InterventionGroup",
"intervention": [
{
"id": "http://data.cochrane.org/interventions/o03rQ3ZMK2h4DJ",
"type": "Intervention",
"childIntervention": [
{
"id": "http://data.cochrane.org/interventions/6nRP2ROypQHa8q",
"type": "Intervention",
"appliedIntervention": {
"id": "http://data.cochrane.org/concepts/r4hp0r2dwmn5",
"type": "Drug",
"label": "Placebo"
},
"interventionClassification": {
"id": "http://data.cochrane.org/concepts/q25gz0m8n54j",
"type": "InterventionClassification",
"label": "No active treatment"
}
},
{
"id": "http://data.cochrane.org/interventions/XMwNewOg1mf3KY",
"type": "Intervention",
"appliedIntervention": {
"id": "http://data.cochrane.org/concepts/r4hp5zcz3yj6",
"type": "Procedure",
"label": "Antibiotic Therapy"
},
"interventionClassification": {
"id": "http://data.cochrane.org/concepts/kn3ptfq7c6lz",
"type": "InterventionClassification",
"label": "Pharmacological Interventions"
}
}
]
},
{
"id": "http://data.cochrane.org/interventions/o0PqJ1yxP5s4AQ",
"type": "Intervention",
"appliedIntervention": {
"id": "http://data.cochrane.org/concepts/r4hp5zcz3yj6",
"type": "Procedure",
"label": "Antibiotic Therapy"
},
"interventionClassification": {
"id": "http://data.cochrane.org/concepts/kn3ptfq7c6lz",
"type": "InterventionClassification",
"label": "Pharmacological Interventions"
}
}
]
},
"outcomeGroup": [
{
"id": "http://data.cochrane.org/outcome-groups/7wqPeqM04dfRP1",
"type": "OutcomeGroup",
"ordinal": "0",
"outcome": {
"id": "http://data.cochrane.org/outcomes/XMxvqDoJxRU3Aw",
"type": "Outcome",
"outcomeClassification": {
"id": "http://data.cochrane.org/concepts/q25g9q497cwk",
"type": "OutcomeClassification",
"label": "Mortality"
},
"outcomeDomain": {
"id": "http://data.cochrane.org/concepts/DAezymWdeqT7y0",
"type": "Condition",
"label": "All Cause Mortality"
},
"outcomeText": "All-cause mortality within 30 days after randomisation"
}
},
{
"id": "http://data.cochrane.org/outcome-groups/E7APMA6pJYue3v",
"type": "OutcomeGroup",
"ordinal": "1",
"outcome": {
"id": "http://data.cochrane.org/outcomes/6n13joJN1KFaOg",
"type": "Outcome",
"outcomeClassification": {
"id": "http://data.cochrane.org/concepts/q25gz0m8n54b",
"type": "OutcomeClassification",
"label": "Device/intervention failure"
},
"outcomeDomain": {
"id": "http://data.cochrane.org/concepts/r4hp3qh9r605",
"type": "Condition",
"label": "Treatment Failure"
},
"outcomeText": "Early clinical failure"
}
}
]
}COVID-19 Study Register Architecture
When the COVID-19 Coronavirus crisis gathered pace, as an organisation specialising in publishing the gold-standard in healthcare evidence, it was clear to Cochrane they needed to leverage their data platform and draw upon all their expertise to aggregate and publish the rapidly growing corpus of clinical evidence being generated globally by researchers and practitioners. The flexibility of the Cochrane linked data platform seemed like the ideal way of making this a reality in quick time.
While the platform until now was primarily focussed on the augmentation of Cochrane reviews, for the new study register we needed to augment Studies and their referenced reports with PICOs prior to going through the Cochrane review process. This required integration with the CRS, the Cochrane Register of Studies. Several steps were needed :
- Integration of the PICO annotation widget into CRS
- Updates to the Content model to support the data structures in the CRS
- ETL of COVID-19 Studies into the knowledge graph (conforming to the Content model)
- Exposing the Studies out via a data service
- Design and build of the COVID-19 application itself
Step (1) was relatively easy, as the PICO annotator was already designed as a widget for dropping into any web based content production system, and for annotating any URI referenceable target content.
Step (2) updating the model. This is where RDF based knowledge graphs excel - in flexible data modelling. Updating our content model with enhanced semantics for Study entities was a doddle. Drawing it up visually first and validating the model with Cochrane subject matter experts. Creating the RDF / Ontology changes for deployment into the graph as a SPARQL update. Deployed using our automated deployment scripts via CI.
Step (3) build a lightweight NodeJS ETL script to ingest studies from the CRS API, transform into RDF, and load into the graph using a SPARQL update. Containerised and deployed using Helm into EKS, scheduled as a Kubernetes batch job.
Step (4) create a new ElasticSearch Connector to materialise the studies into a new elastic index, and expose via the existing Content API.
Step (5) is where it got even more interesting. We wanted to be able to rapidly build a completely new web app for searching and navigating Studies using PICOs. Building with ReactJS let us re-use components from other Cochrane apps we had already developed, and to aid data binding and state management we opted for GraphQL, specifically Amazon’s AWS AppSync GraphQL service.
The conceptual architecture for the COVID-19 Study Register looks like this :
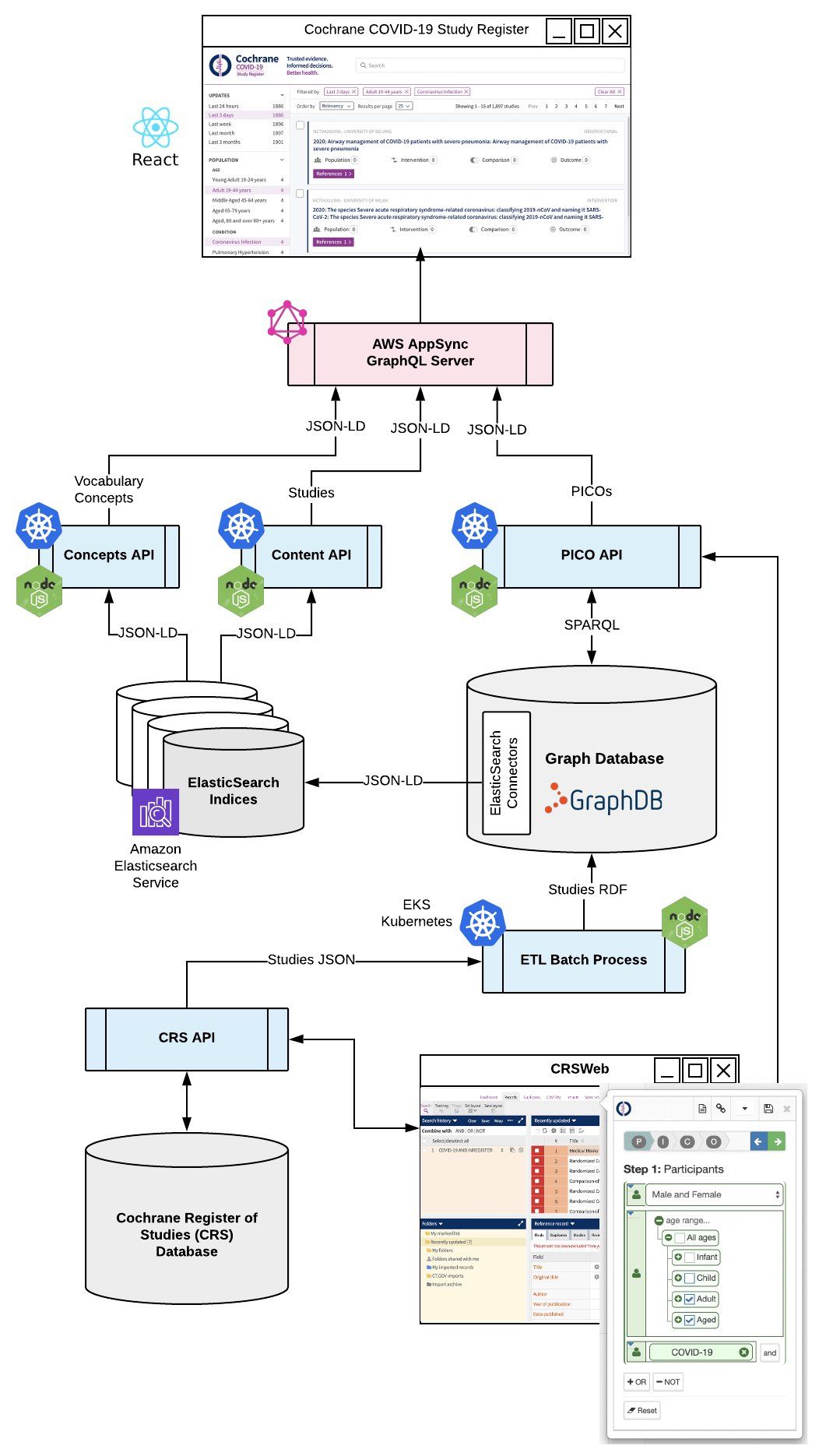
GraphQL
We chose to use GraphQL having used it very successfully on another project earlier this year, and primarily for these reasons :
- Rapid application development - the AppSync / Apollo server implementation works beautifully with React - integrating with React Hooks for query and subscriptions. In-browser developer tools akin to the redux-devtools for GraphQL debugging, and you can manage local app state using the GraphQL schemas and queries.
- Sharing of ontology and schemas: the GraphQL schemas align nicely with the ontology models that we describe our data with, whether that data is within the knowledge graph or as messages between services.
- The ability for us to collate data and dereference data from a number of data sources server-side, allowing the client-side app to use only what it needs, to reduce chatter.
Going forwards, it would be great to explore the use of GraphDB’s new GraphQL API with the cochrane data.
Conclusive Evidence
When I originally started writing this blog post some weeks back, it was going to be about the history behind the Cochrane Linked Data Platform, and how Cochrane have successfully used knowledge graph technology to change the way they think about data, and how linked data has become one of the central pillars of their data strategy. However, having now worked hard and fast with our team and the Cochrane team over the last two weeks to build a COVID-19 Study Register at breakneck speed on top of the platform, it made a much more compelling story, to talk about how we achieved this.
It has led me to conclude several key takeaways :
- It has demonstrated the supreme flexibility in ontology driven data architectures for rapid adaptation and evolvability
- Bounded context domain and ontology modelling patterns provide clean delineation between data concerns, allowing you to confidently adapt parts of your data landscape without leaking potentially breaking changes into other parts.
- Don’t take shortcuts with respect to deployment, infrastructure and test automation when building your systems - it pays large dividends down the road when you need your platform to evolve.
- If you think you can't build extremely robust, scalable platforms efficiently using linked data and knowledge graph technologies, think again.

